The Ellgaard Lab
We aim to uncover structure, function and evolution of conotoxins – disulfide-rich venom peptides and proteins from predatory marine cone snails. To aid this work, we also develop E. coli expression systems for disulfide-rich proteins, which allows us to explore new families of conotoxins and their potential application as research tools and drug leads.
Predatory marine cone snails are amazing creatures that kill their prey – fish, worms or other snails – by injecting them with a deadly mixture of hundreds of different disulfide-rich neuropeptides referred to as conotoxins. Conotoxins are highly selective compounds that can target specific subtypes of receptors or ion channels often located in the nervous system, a characteristic that has led to their wide use in ion channel research and as therapeutic agents. The fact that conotoxins generally contain many disulfide-bonds make them hard to produce synthetically (at least without prior knowledge of their disulfide pattern). To overcome this bottleneck, we have designed expression systems for production of disulfide-rich proteins in the cytosol of E. coli, in part relying on co-expression with the cone snail enzymes that catalyze disulfide-bond formation in venom gland cells.
Our main focus is the characterization of conotoxins belonging to newly discovered superfamilies – toxins without known structure or function. We are particularly interested in understanding the connection between sequence, structure, and function of large conotoxins (aka macro-conotoxins). In addition, we have ongoing projects that focus on establishing a method for unbiased identification of conotoxin targets and on protein folding and misfolding in the ER of mammalian cells (a long-standing area of research in the lab).
Characterization of conotoxins belonging to new superfamilies
The advent of new sequencing technologies and bioinformatics tools for transcriptome analysis has revealed thousands of previously unknown animal venom peptide and protein sequences in recent years. Data mining of venom gland transcriptomes has revealed a large number of new conotoxin superfamilies. Informed by AlphaFold structure prediction of such sequences we have selected a number of macro-conotoxins for detailed characterization. Our experience tells us that each new conotoxin provides fascinating insight into sequence/structure/function relations – and that every new toxin we work on is full of surprises! With this work we aim to connect the biochemical and molecular characteristics of macro-conotoxins with the biology and behavior of cone snails. These studies are done in close collaboration with the labs of Profs. Baldomero “Toto” Olivera and Helena Safavi-Hemami, University of Utah, USA, which are experts on venom protein evolution and have pioneered assays to study conotoxin function on neuronal cells.
- Hackney, C.M. et al.,
- Nielsen, L.D. et al., J. Biol. Chem. (2019), 294, 8745-8759.
- Safavi-Hemami, H. et al., PNAS (2015), 112, 1743-1748.
Venom peptide production
To accelerate venom peptide discovery, we have developed two expression systems, csCyDisCo (based on the original CyDisCo system from the Ruddock lab, University of Oulu, Finland) and a more recent version called DisCoTune, that allow the production of highly disulfide-bonded proteins in the cytosol of E. coli. Currently, we are establishing a high-throughput synthetic biology platform (DisCoTech) that will allow efficient screening of thousands of sequences encoding disulfide-rich proteins of biomedical interest to quickly identify those that produce well in the DisCoTune system. These efforts are conducted together with Senior Researcher Morten Nørholm, Technical University of Denmark.
- Rivera-de-Torre, E. et al., Front. Bioeng. Biotechnol. (2022), 9, 811905.
- Bertelsen, A.B. et al., Microb. Biotechnol. (2021), 14, 2566–2580.
- Nielsen, L.D. et al., J. Biol. Chem. (2019), 294, 8745-8759.
- Safavi-Hemami, H. et al., PNAS (2016), 113, 3227-3232.
Identifying venom peptide targets
With the ability to produce many conotoxins recombinantly, a next challenge is the identification of their endogenous molecular targets. Using a variety of toxins from spider, scorpion, and cone snail with both known and unknown targets, we are aiming to establish an unbiased method for target identification. This work is a collaboration with Dr. Rocio Finol-Urdaneta, University of Wollongong, Australia, and Assoc. Prof. Helena Safavi-Hemami, University of Utah, USA.
Conotoxin structure determination
An integral part of the characterization of new conotoxins is their structure determination by either NMR spectroscopy (in collaboration with Prof. Kaare Teilum, University of Copenhagen) or X-ray crystallography (in collaboration with Prof. Preben Morth, Technical University of Denmark). Here are two recent examples – with more on the way soon:
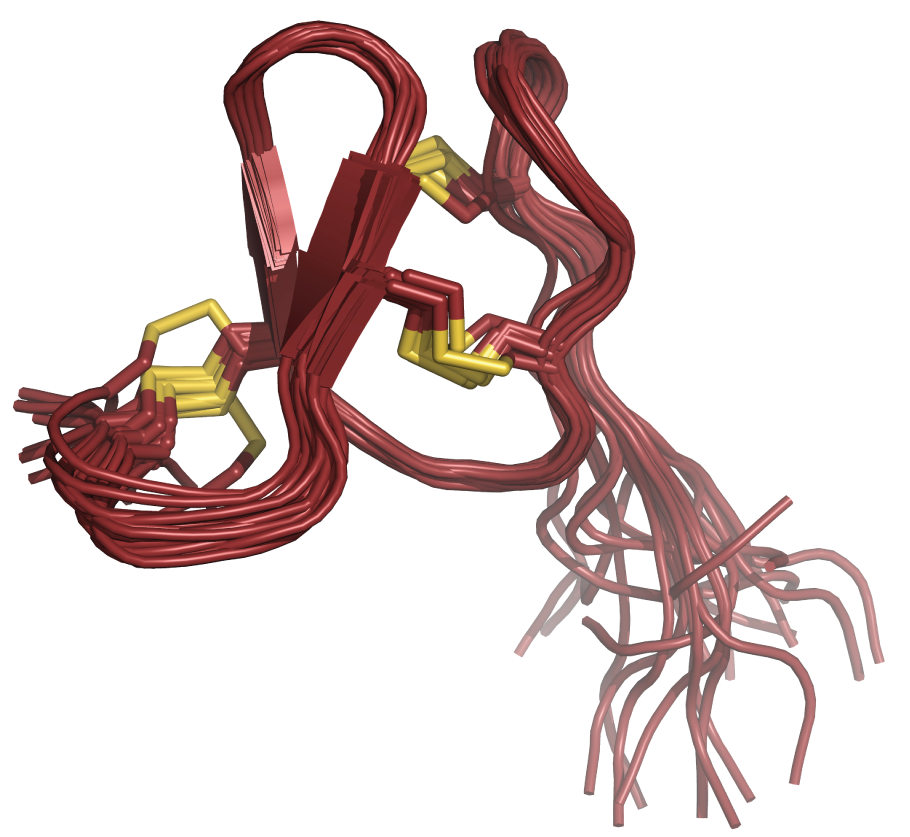 |
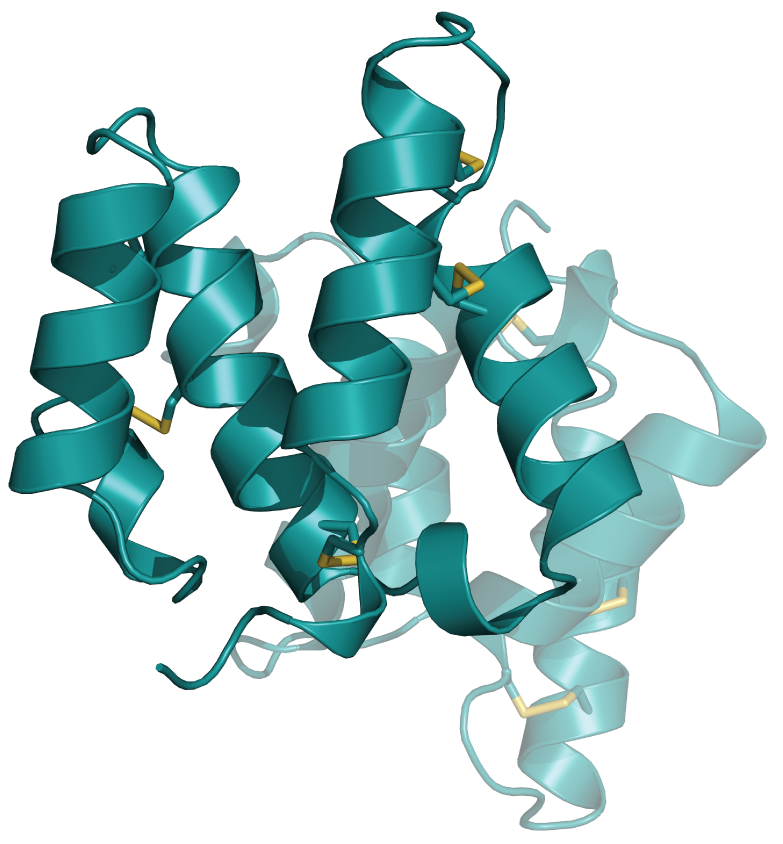 |
| H-Vc7.2 from Conus vitoriae | Mu8.1 from Conus mucronatus |
Protein folding and misfolding in the ER
About a third of all mammalian proteins are synthesized in the ER. Throughout the years we have studied how these proteins fold, with a particular focus on enzymes that catalyze disulfide-bond formation, and what happens when they misfold and are degraded (a process termed ER-associated degradation). Current work in collaboration with Assoc. Prof. Lina Dahlberg, Western Washington University, USA, investigates the function of a specific component of the ERAD machinery in C. elegans.
We are always happy to receive applications from potential postdocs, Ph.D., Masters, Erasmus and Bachelor students and welcome applications from both Denmark and abroad. If you are interested in joining the lab or just want to know more about our work, contact Lars by phone or email – or stop by the lab. Our group has close scientific (and social) interactions with the rest of the Protein Biology Group, the Section for Biomolecular Sciences and the Kaj Ulrik Linderstrøm-Lang Centre for Protein Science.
Postdoc positions
Currently we have no open funded positions, but the chances of obtaining postdoc grants are quite good when applying with Danish and/or international foundations.
PhD projects
The University of Copenhagen occasionally offers studentships for students holding a Danish or foreign Masters degree. In addition, possibilities exist to apply for funding from private foundations. Phd students are accepted into the PhD school at the Faculty of Science.
Masters, Bachelor and Erasmus student projects
We always offer Masters, Bachelor and Erasmus projects within our different areas of research. For more information just drop me a line. You can also find information about these types of projects by clicking on the "Ellgaard student projects"-link on the right.
For all types of positions, please contact Lars concerning availability and to discuss specific projects.
- Welcome to Enriko who joins the lab as a PhD student (1.12.2023)
- Two papers out on the structure, function and evolution of the Mu8.1 macro-conotoxin from Conus mucronatus (August 2023)
We are grateful for current funding from the Novo Nordisk Foundation and the Independent Research Fund Denmark (FNU).
Our past research was funded by the Novo Nordisk Foundation, Independent Research Fund Denmark (FNU and FTP), Lundbeck Foundation, Benzon Foundation, Carlsberg Foundation, European Commission, Villum Foundation, A.P. Møllers Fond til Lægevidenskabens Fremme, Augustinus Fonden, Foundation JUCHUM, Brdr. Hartmanns Fond, Ulla og Mogens Folmer Andersens Fond, Hørslev-Fonden, Lizzi og Mogens Staal Fonden, Swiss National Science Foundation, ETH Zürich Research Commission, Roche Research Foundation, and Novartis Stiftung.
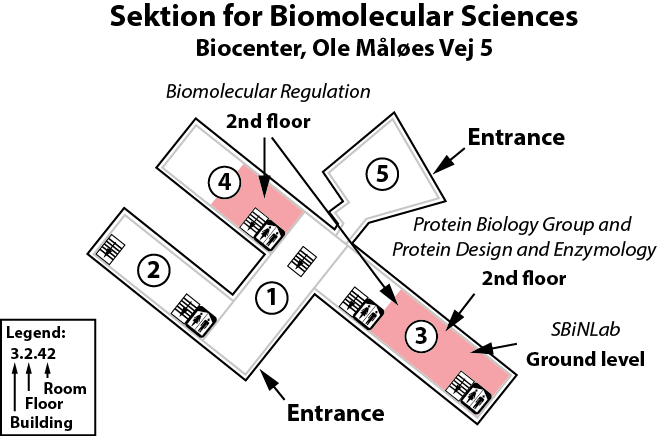
The Ellgaard lab is located on the 2nd floor, building 3 of the Copenhagen Biocenter (map), close to the centre of Copenhagen. It takes less than 1 hour to travel from Copenhagen's airport to our lab, by train or subway (from the airport to Nørreport station, a 20 minute journey, with trains every 10 minutes) and bus (bus 150S from Nørreport to the stop called "Fredrik Bajers Plads" in the intersection of Tagensvej and Nørre Allé, a 10 minute journey, with buses every 6-8 minutes). From there it is a few minutes walk.
Postdocs
• Saad Khilji (2022-)
• Celeste Hackney (2022-2023)
Now Lab Manager in our group
• Lau Dalby Nielsen/Kjelgaard (2017-2020)
Research scientist at Chr. Hansen
• Giang Nguyen (2018)
Postdoc in the lab of Andreas Laustsen, DTU
• Helena Safavi-Hemami (2013-2017)
Associate Professor, University of Utah
• Raffaela Magnoni (2013-2015)
Senior Scientist at Orphazyme, Copenhagen
• Andrea Vala (2007-2010)
Senio Scientist at Symphogen
• Christian Appenzeller-Herzog (2004-2009)
Scientific Librarian at University of Basel, Switzerland
• Linda Johansson (2006-2008)
Regulatory Affairs Manager at Astra Zeneca, Mölndal, Sweden.
Ph.D. students
• Enriko Robin Raidjõe (2023-)
• Celeste Hackney (2018-2022)
Now Lab Manager in our group
• Minttu Virolainen (2016-2020):
Medical writer at ALK
• Ana Paula Cordeiro (2014-2018):
Protein and Formulation Scientist at Evaxion Biotech
• Lea Cecilie Christensen (2011-2016):
Project employee at Astra
• Njal Winther Jensen (2011-2014):
Employed at Vifor Pharma
• David Månsson (2010-2013):
Upstream specialist at Lundbeck
• Henning Gram Hansen (2009-2013):
Senior scientist at Novo Nordisk
• David Klinkenberg (2012-2013):
Bioproduction Scientist at Thermo Fisher Scientific
• Jan Riemer (2003-2008):
Professor, University of Cologne.
• Doris Roth (2003-2008):
Patent Consultant at Guardian IP Consulting, Copenhagen
• Johannes Haugstetter (2002-2007):
Scientist at InSphero, Zurich, Switzerland.
Special guests
• Lina Dahlberg, visiting professor (2021):
Associate professor, Western Washington University, USA
• Amie McClellan, visiting professor (2018):
Faculty, Bennington College, USA
• Eva Frickel, Ph.D. student & postdoc (2000-2005):
Senior Wellcome Trust Fellow, Reader, University of Birmingham, UK
• Christiane Schirra, technician (2001-2003)
• Manuel Fischer, Ph.D. student (2013):
"On loan" three months from the Riemer lab
Research assistants
• Nicklas Lund Ryding (2023-24)
Studies Biochemistry, UCPH
• Ellen Johanne Olsen (2020-21)
Employed at Bioneer
• Sophie Heiden Laugesen (2019)
PhD student with Helena Safavi-Hemami, UCPH
• Jurate Kamarauskaite (2010-2011)
Did her PhD with Peter Kast, ETH Zürich
Master's students
• Leonardo Marcazzan (2023)
• Camilla Høgfeldt Jessen (2021-22)
• Mikkel Dyring Andersen (2021-22)
Employed at Novo Nordisk
• Camilla Skotte Pedersen (2021)
• Josefine Rahbek (2021)
Employed at Expres2ion Biotechnologies
• Karoline Leth (2020)
Clinical Research Associate hos IQVIA
• Anna Damsbo Jensen (2019)
Research Scientist at Bactolife (PhD with Andreas Laustsen, DTU)
• Queralt Ginebra (2019)
Studied Molecular Biomedicine, UCPH
• Sophie Heiden Laugesen (2018)
PhD student with Helena Safavi-Hemami, UCPH
• Andreas Birk Bertelsen (2018)
PhD student with Senior Researcher Morten Nørholm, DTU
• Emilie Müller (2017)
Protein and Downstream Process Scientist at 21st.BIO (PhD with Prof. Preben Morth, DTU)
• Celeste Hackney (2017)
Now lab Manager in our group (also did her PhD in our lab)
• Sara Ambjørn (2016)
Postdoc with Jakob Nilsson, UCPH (PhD with Vibe Østergaard, UCPH)
• Mads M. Foged (2016)
Postdoc at the University of Lausanne (PhD with Kenji Maeda, Danish Cancer Society Research Center)
• Minttu Virolainen (2016)
Medical writer at ALK (also did her PhD in our lab)
• Mark Oglesby (2015)
Technical Operations Specialist, William Cook Europe, LifeScience Services
• Simon Kamenov (2015)
Research Scientist (mass spectrometry), Novo Nordisk (performed an industrial PhD at Novo and Aalborg University)
• Line Louise Stolze Larsen (2014)
Project Manager, Novo Nordisk
• Sofie Engfred Larsen (2014)
Microbiologist and GMP coordinator, Novo Nordisk
• Carsten S. Nielsen (2012)
Development Scientist, Novo Nordisk (PhD with Peter Brodersen, UCPH)
• Sidsel Nag (2012)
Senior Experience Strategist, Halfspace (PhD at HEALTH, UCPH)
• Lea C. Christensen (2011)
Project employee at Astra (PhD in our lab)
• Cecile Lützen Søltoft (2011)
QA Assistant at Nordic Bioscience
• Jonas Damgård Schmidt (2010)
Senior scientist at Novo Nordisk (PhD at the Panum Institute, UCPH)
• Njal Winther Jensen (2009)
Employed at Vifor Pharma (did his PhD in our lab)
• Nils Althaus (2005):
Actor and singer-songwriter - check out his homepage.
• Michael "Miguel" Maurer (2005):
Principal Scientist at novaGo Therapeutics, Switzerland (Ph.D. at the University of Zürich).
• Jaco den Otter (2005)
• Annika Kyburz (2004):
R&D Scientist at Thermo Fisher Scientific, Helsinki, Finland (Ph.D. with Ilkka Kilpeläinen, University of Helsinki).
• Karl Bihlmaier (2004):
Medical Doctor at Universitätsklinikum Erlangen, Germany (Ph.D. with Johannes Herrmann, University of Kaiserslautern).
• Micha Häuptle(2003):
Senior Director / Head of CMC at Vector BioPharma Basel, Switzerland (Ph.D. with Thierry Hennet, University of Zürich)
• Vital Wohlgensinger (2002):
Chemistry teacher at ISME St. Gallen und Sargans (Ph.D. student with Reinhardt Seger, Kinderspital Zürich)
• Anna Söderholm (2002):
Ph.D. with Heribert Stoiber, University of Innsbruck.
• Susanne Sales (1999):
Ph.D. with Peter Sonderegger, University of Zürich.
Technicians and Lab Managers
• Cecile Lützen Søltoft (2013-2023)
QA Assistant at Nordic Bioscience
• Kelli Louise Doherty (2012-2013)
Senior Laboratory Technician at Novo Nordisk
• Zhila Nikrozi (2008-2012):
Now at the Core Facility for Integrated Microscopy, University of Copenhagen.
• Sandra Abel Nielsen (2007):
Senior Scientist at Shennon Biotechnologies, San Francisco, USA
• Christel Elkjær Olsen (2006-2007)
Now a landscape gardener
• Martin Pagac (2003-2005):
Senior Scientist at dsm-firmenich (Ph.D. with Andreas Conzelmann, University of Fribourg).
Technician trainees
• Morten Bjerre Nielsen (2007-2008):
Completed a bachelor degree in biology. Now technician at the University of Copenhagen.
• Caroline Ashiono (2004-2005):
Technician with Ulrike Kutay, ETH Zürich.
• Gwynneth Zimmermann (2003-2004):
Technician with the therapeutic antibody company Esbatech, Zürich.
 December 2023: Johanna Richter, Oliver Kønig-Wheatley, Nicklas Lund Ryding, Saad Khilji, Celeste M. Hackney, Lars, Alexander Diatchikhine and Enriko Raidjõe.
December 2023: Johanna Richter, Oliver Kønig-Wheatley, Nicklas Lund Ryding, Saad Khilji, Celeste M. Hackney, Lars, Alexander Diatchikhine and Enriko Raidjõe.
Nov 2018: Cecilie L. Søltoft, Celeste M. Hackney, Minttu Virolainen, Lau D. Nielsen/Kjelgaard, Lars, Anna D. Jensen and Jens H. Sørensen.

December 2016: Mia Øgaard, Celeste M. Hackney, Cecilie L. Søltoft, Helena Safavi-Hemami, Lars, Ana P. Cordeiro, Mads M. Foged, Emilie Müller, Minttu Virolainen and Simona Kotesova.

May 2015: Back - Sara M. Ambjørn, Cecilie L. Søltoft, Lars, Mark Oglesby, Simon Kamenov. Front - Raffa Magnoni, Ana P. Cordeiro, Lea C. Christensen, Minttu Virolainen.
 December 2013: Back - Cecilie L. Søltoft, Raffa Magnoni, Lars, Mark Oglesby, Sofie E. Larsen. Front - Manuel Fischer, Njal W. Jensen, Lea C. Christensen, Line L.S. Larsen.
December 2013: Back - Cecilie L. Søltoft, Raffa Magnoni, Lars, Mark Oglesby, Sofie E. Larsen. Front - Manuel Fischer, Njal W. Jensen, Lea C. Christensen, Line L.S. Larsen.
 December 2012: Kelli Doherty, Niels B. Holm, David Månsson, Njal W. Jensen, Lars, Sofie E. Larsen, Henning G. Hansen, Lea C. Christensen, Oliver Krigslund, Line L.S. Larsen, David Klinkenberg.
December 2012: Kelli Doherty, Niels B. Holm, David Månsson, Njal W. Jensen, Lars, Sofie E. Larsen, Henning G. Hansen, Lea C. Christensen, Oliver Krigslund, Line L.S. Larsen, David Klinkenberg.
 April 2011: Back - Henning G. Hansen, Lars, Sidsel Nag, Carsten S. Nielsen, Cecilie L. Søltoft, Njal W. Jensen, Lea C. Christensen, David Månsson. Front - Jurate Kamarauskaite, Zhila Nikrozi.
April 2011: Back - Henning G. Hansen, Lars, Sidsel Nag, Carsten S. Nielsen, Cecilie L. Søltoft, Njal W. Jensen, Lea C. Christensen, David Månsson. Front - Jurate Kamarauskaite, Zhila Nikrozi.
 June 2009: Back - Njal W. Jensen, Henning G. Hansen, Andrea Vala, Lars, Jonas D. Schmidt. Front - Christina Brandt, Sine Godiksen, Zhila Nikrozi.
June 2009: Back - Njal W. Jensen, Henning G. Hansen, Andrea Vala, Lars, Jonas D. Schmidt. Front - Christina Brandt, Sine Godiksen, Zhila Nikrozi.
 June 2008: Rikke Rasmussen, Cecilie, L. Søltoft, Zhila Nikrozi, Doris Roth, Jan Riemer, Christian Appenzeller-Herzog, Sine Godiksen, Lars.
June 2008: Rikke Rasmussen, Cecilie, L. Søltoft, Zhila Nikrozi, Doris Roth, Jan Riemer, Christian Appenzeller-Herzog, Sine Godiksen, Lars.
 February 2006: Martin Pagac, Johannes Haugstetter, Niels Althaus, Christian Appenzeller-Herzog, Caroline Ashiono, Doris Roth, Lars, Michael Maurer, Jan Riemer.
February 2006: Martin Pagac, Johannes Haugstetter, Niels Althaus, Christian Appenzeller-Herzog, Caroline Ashiono, Doris Roth, Lars, Michael Maurer, Jan Riemer.

Contact

Professor Lars Ellgaard
Section for Biomolecular Sciences
Ole Maaløes Vej 5, room 3-2-31
DK-2200 Copenhagen N
Email: lellgaard_at_bio.ku.dk
Phone: +45 3532 1725
KU profile: Link